Non-redundant TF motif matches genome-wide
Last updated:
Overview
The redundancy and overlap between various motif model databases complicate downstream analysis and interpretation. We computed the pairwise similarity for >2,000 motif models determined for both human and mouse TFs and clustered them into 286 distinct motif clusters. Within each cluster we aligned motifs to each other to generate an archetypal consensus motif.
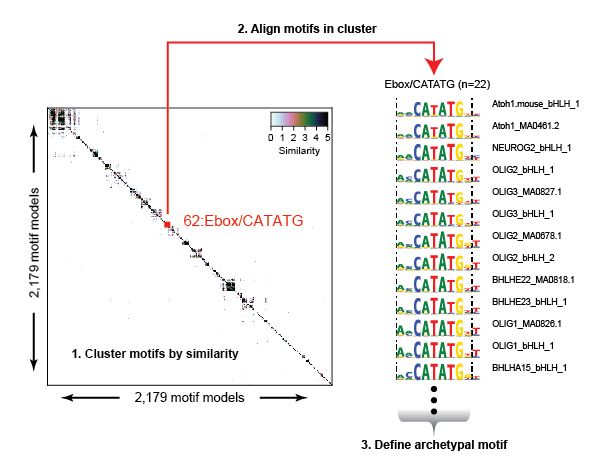
We scanned the entire genome using each motif model (all 2,179 models), and then repositioned the coordinates of each genomic match according to each motifs relative position to its archetypal model. After adjusting the coordinates of each motif match, we remove all duplicates (same position and motif cluster), retaining a single match at any position in the genome.
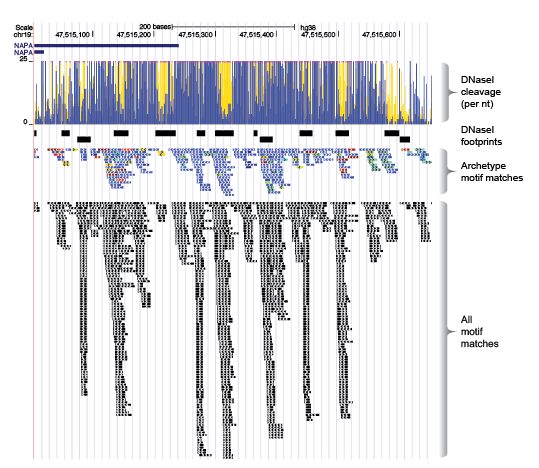
Some key advantages of the this approach:
- Allow sharing of information about TFBS from many models. Any match to an individual model is a match to the cluster
- No information is lost about individual motif model matches. The individual motif matches are fully represented in each cluster archetype match
- Agnostically identifies errors and mislabeling within motif databases
Some notable caveats/shortcomings that should be considered:
- No optimization on the bit-score/p-value threshold for each motif model
- Cluster tree is cut at an abitrary height (chosen by the look and feel of the clusters)
Included motif databases (v1.0)
JASPAR 2018: Models derived from published and experimentally defined transcription factor binding sites for eukaryotes. The “CORE vertabrates” non-redundant PFMs were included.
- Motif models [MEME-format]
- Motif weblogos [EPS-format]
Taipale 2013 HT-SELEX: Models derived from high-throughput SELEX experiments using bacterially expressed human and mouse DNA-binding domains. See Table S3 from publication.
- Motif models [MEME-format]
- Motif weblogos [EPS-format]
HOCOMOCO version 11: Models derived from ChIP-seq data (680 human + 452 mouse TFs)
- Motif models [Human: MEME-format, Mouse: MEME-format]
- Motif weblogos: [EPS-format]
Cluster visualization
Visit this page to view the 286 clustered motif models (v1.0).
Browser tracks
To visualize the motif matches in the genome, I have created a trackhub for the UCSC Genome Browser. You can load this track by navigating on any UCSC browser instance to “Data → Trackhubs” and then copying and pasting the following URL:
https://resources.altius.org/~jvierstra/projects/motif-clustering/releases/v1.0/hub.txt
Alternatively, you can click here to automatically load this trackhub at the Genome Browser hosted at UCSC.
Downloads
v1.0 (2020-4-14)
Clustering of 2179 motif models (3 databases above)
- Motif clustering annotations (excel file)
-
Genome-wide motif scans
Genome build Full scans Archetype scans Human (hg38) bed.gz + tabix, bigBed bed.gz + tabix, bigBed Mouse (mm10) bed.gz + tabix, bigBed bed.gz + tabix, bigBed Note: Theses files are massive (>~40Gb). We have provided a TABIX-index alongside to facilitate remote access using
tabix:[jvierstra@test0 $] tabix https://resources.altius.org/~jvierstra/projects/motif-clustering/releases/v1.0/hg38.archetype_motifs.v1.0.bed.gz chr19:45,001,882-45,002,279 chr19 45001871 45001883 ZNF306 4.4101 - ZNF306_C2H2_1 1 chr19 45001891 45001907 TBX/1 7.8289 + BRAC_HUMAN.H11MO.0.A 1 chr19 45001896 45001916 GC-tract 8.4712 + ZN467_HUMAN.H11MO.0.C 1 chr19 45001897 45001914 KLF/SP/2 13.2278 + PATZ1_HUMAN.H11MO.0.C 5 chr19 45001898 45001908 HD/12 8.7818 + PBX3_MA1114.1 1 ...
File format descriptions
Full scans
| Column | Example | Description | |
|---|---|---|---|
| 1 | contig |
chr1 | Chromosome |
| 2 | start |
10003 | Start position (0-based) |
| 3 | stop |
10023 | End position |
| 4 | motif |
RREB1_MA0073.1 | Motif model name |
| 5 | match_score |
3.75359698976 | MOODS match score |
| 6 | strand |
+ | Matching strand |
| 7 | seq |
CCCTAACCCTAACCCTAACC | Genomic sequence of match |
Archetype scans
| Column | Example | Description | |
|---|---|---|---|
| 1 | contig |
chr1 | Chromosome |
| 2 | start |
10005 | Start position (0-based) |
| 3 | stop |
10022 | End position |
| 4 | motif_cluster |
KLF/SP/2 | Motif cluster name |
| 5 | match_score |
3.7536 | MOODS match score for best cluster match |
| 6 | strand |
+ | Matching strand |
| 7 | best_model |
RREB1_MA0073.1 | Best matching motif model from cluster |
| 8 | num_models |
1 | Number of motif model from cluster with a match |
Code
Software and scripts are available at GitHub to reproduce the motif clustering.
Notes
Motifs scans were performed with MOODS
python2 moods_dna.py \
--sep ";" -s h38.fa --p-value 0.0001 \
--lo-bg 2.977e-01 2.023e-01 2.023e-01 2.977e-01 \
-m ${PFM_FILE} -o ${OUTFILE}
Citation
If you use this resource in your research, please kindly cite:
Vierstra2020 Vierstra, J., Lazar, J., Sandstrom, R. et al. Global reference mapping of human transcription factor footprints. Nature 583, 729–736 (2020)
Additional resources
Admittedly, I am far from the first person to cluster motif models. In any case, this resource is/was largely intended to aid in visualizing and “surfing” the genome browser and to accompany other annotations (e.g., ChIP-seq, DGF, etc.).
Below I have compiled a list of publications, websites and databases pertaining to the curation of motif models which may of be of use. If you find something useful or if I have missed something, please contact me and I will include it here.
Clustering:
- RSAT: Clustering methodology used as part of JASPAR project (publication link)
- TOMTOM: Software tools used here for clustering (publication link)
Motif curation:
- JASPAR
- HOCOMOCO
- CisBP (Tim Hughes’ group)
- Lambert et al.: “The human transcription factors” (Supplementary Table 1)
- UniProbe (Martha Bulyk)